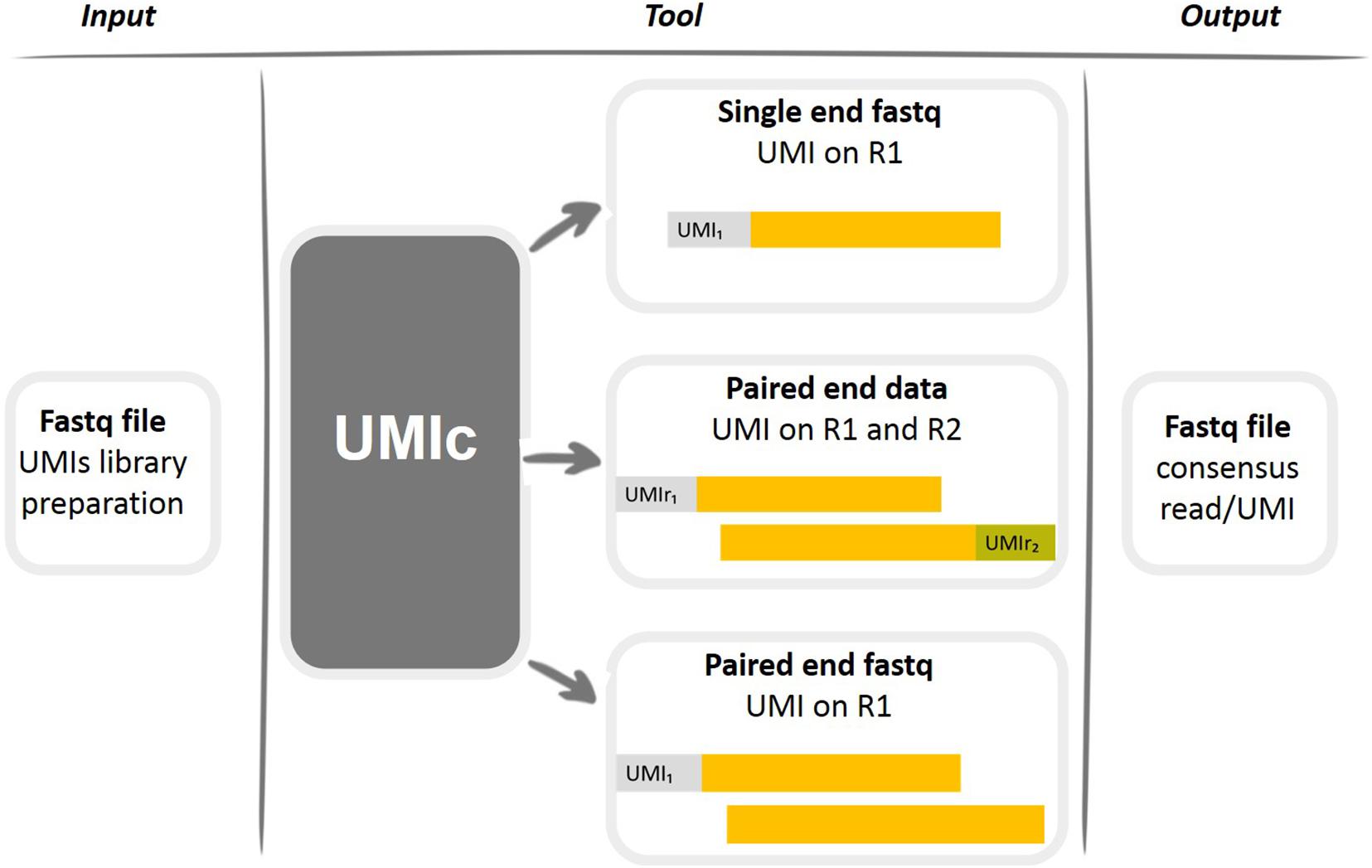
UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv

Comparative analysis of NovaSeq 6000 and MGISEQ 2000 single‐cell RNA sequencing data - Chen - 2022 - Quantitative Biology - Wiley Online Library

UMI-Gen: A UMI-based read simulator for variant calling evaluation in paired-end sequencing NGS libraries - ScienceDirect

Effects of pre-processing pipelines on the number of genes detected... | Download Scientific Diagram

Analysis of UMIs. a Stacked bar plot showing the fractions of unique... | Download Scientific Diagram

UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv
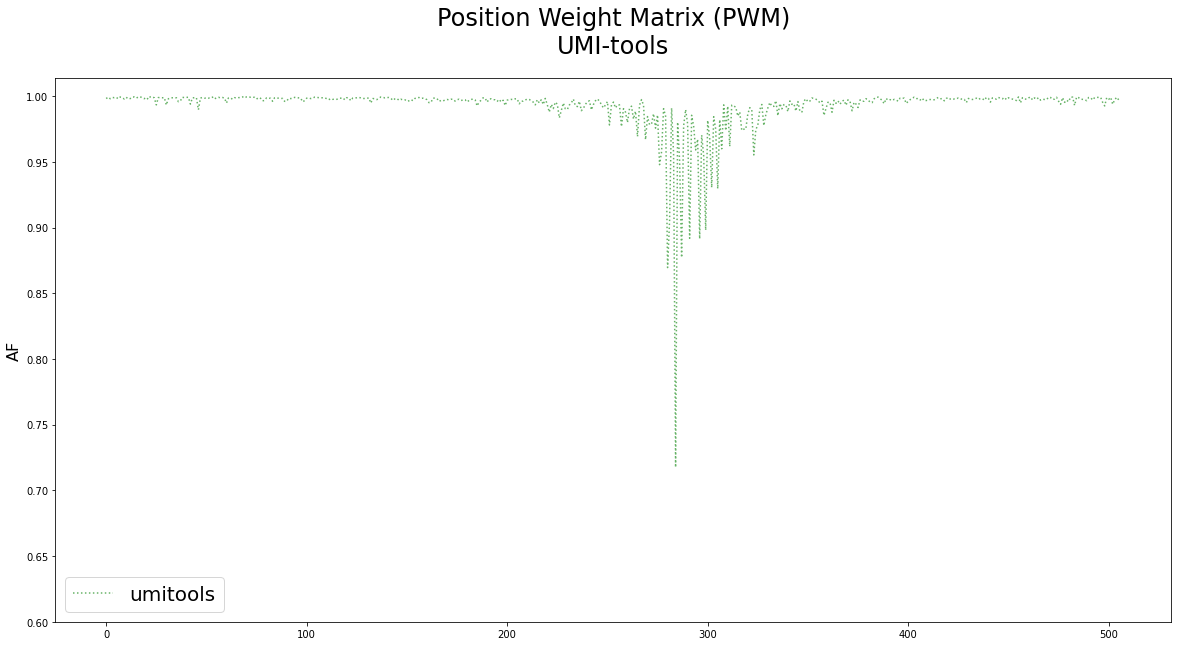
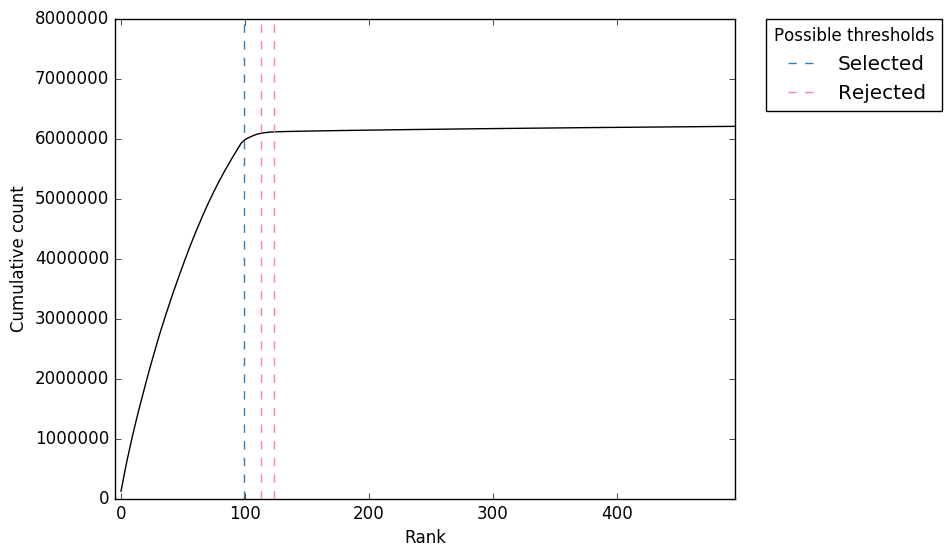
![PDF] Title UMI-tools : Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy Running | Semantic Scholar PDF] Title UMI-tools : Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy Running | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/1b184f51965099c10ef44c76d7d1572184fb678d/16-Figure4-1.png)
PDF] Title UMI-tools : Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy Running | Semantic Scholar